ISSN 2410-5708 / e-ISSN 2313-7215
Year 8 | No. 21 | p. 94 - p. 102 | February - May 2019
© Copyright (2019). National Autonomous University of Nicaragua, Managua.
This document is under a Creative Commons
Attribution-NonCommercial-NoDerivs 4.0 International licence.
Carbapenemase-producing Pseudomonas aeruginosa from Managua, Nicaraguan hospitals
https://doi.org/10.5377/torreon.v8i21.8851
Submitted on Octuber 3rd, 2019 / Accepted on October 30th, 2019
M.SC. Oscar Arbizú Medina
Molecular Biology Laboratory “MA. Elmer Cisneros in memoriam”, Polytechnic Institute of Health
UNAN-Managua
arbizu2006@yahoo.com.mx
BA. Kenia García Rosales, BA. Benjamín Castillo Gómez, BA. Arelys Mejía Alvares , Ph.D. Abraham Salinas
Collaborating authors
Keywords: Pseudomonas, resistance, carbapenemasa.
Abstract
The current study was presented to know the frequency of Pseudomonas aeruginosa metallo-β-lactamasas and resistance patterns in hospitalized patients. For this, a cross-sectional study was carried out in the period of July 2016, January 2017. The strains were obtained from the different infectious processes of hospitalized patients, strains were preserved in milk broth in hospitals until their subsequent analysis, the strains were reactivated in MacConkey agar, a screening was carried out by the method of Kirby Bauer, the cut-off value was taken, when Imipenem and Meropenem was ≤21mm, the synergy test was determined, using carbapenemics and their inhibitors, ethylenediaminetetraacetic acid (10µg) and phenyl boronic acid (10 µg), the resistance profile was performed using vitek2 Compact, the minimum inhibitory concentration was determined for Imipenem 2-4 µg/ml and for Ertapenem 2 µg/ml, genotyping was determined by Reaction in Polymerase chain and molecular weight was determined by electrophoresis.
The universe was comprised of 288 strains, of these 22 were Pseudomonas aeruginosa, which corresponded to 7.6%, the strains analyzed presented multi-resistance to the various antibiotics analyzed, only 7 (32%), resistance to tigecycline and none of the strains present resistance to Colistin, the most frequently genes was VIM, for 68%, the most frequently type of sample, where Pseudomonas was isolated, was tracheal samples, 7 (32%), followed by secretion 5 (23%), Blood culture 1 (4%), catheter tip 4 (18%), and the most frequent rooms where these microorganisms were present, was a pediatric ICU with 91%.
As a result of the study, it is concluded that the prevalence of Pseudomonas aeruginosa, producers of carbapenemases, in hospitals in Nicaragua, is low, but should be of concern for hospital authorities, because the strains contain various types of genes combined as; VIM and GIM, VIM and SPM, which can trigger epidemic outbreaks, the frequency with which new strains resistant to carbapenems appears is evident, this evidence involves the search for strategies, to avoid the dispersion of multiresistant strains in different health units at the time of transfer of a carrier patient to another hospital unit.
Introduction
Pseudomona aeruginosa, is one of the opportunistic pathogens that causes nosocomial infections, has the ability to colonize the patient’s moisture surface such as the ear, the patient’s airways subjected to mechanical ventilators, catheters tip and inanimate surfaces such as sinks, microorganisms have developed them differently resistance mechanism to survive the constant attacks of antibiotics, this has led to a drastic reduction of therapeutic options to treat some infectious processes, highlighting the risk of life, because it prolongs the stay in hospitals, greater expenditure of resources To treat the infectious process.
In recent years, reports of microorganisms resistant to various antibiotics are increasingly frequent given by various mechanisms that range from the natural as the AMP-C type, and those acquired by mobile genetic elements mainly by plasmids or integrons, enzymes capable of hydrolyzing State-of-the-art antibiotics such as carbapenemases enzymes capable of hydrolyzing carbapenems. Resistance by hydrolytic enzymes is the most relevant because they can be transferred from one microorganism to another, due to the horizontal transfer mechanism that can affect not only gender, but also another very close group that can share similar characteristics (M. Bodi, 2007 ) (Karen Bush, 2010).
Carbapenemic resistance in our hospitals is more frequently due to carbapenemases enzymes capable of hydrolyzing carbapenemics such as β-lactams, these can be classified in class A, B, C and D. Class A, C and D, Serine type β-lactamases include this because of their active site, while types B correspond to metallo-β-lactamases (MBL), dependent on their active site which is zinc; which are divided into three subclasses, subclass B1, groups the inducible enzymes associated with plasmids (IMP, VIM, SPM, GIM, and SIM), being the most common found in this type of microorganism and also frequently distributed in the Americancontinent. Subclasses B2 and B3 group the chromosomal metallo-beta-lactamases derived from Aeromonas spp, Serratia fonticula, and Stenotrophomona smaltophilia. These enzymes are characterized by providing resistance to beta-lactam antibiotics which involve cephalosporins, monobactams and carbapenems excluding Colistin and Tigecycline. MBLs are resistant to Aztreonam, because they bind with very low affinity to monobactams because of the position of the active site that does not favor hydrolysis. The detection of this enzyme, can be detected by the Kirby Bauer method, using sensidisk of ethylenediaminetetraacetic acid (EDTA) 372 µg or Mercapto sodium acetate (SMA) 900 µg, is a cheap method to guide the present resistance mechanism (Armando Guevara, 2009) (Karen Bush, 2010).
Methods
A cross-sectional study was carried out with the objective of knowing the frequency of Pseudomona auriginosa, producer of ba-lactamase type carbapenemases, in which they studied 288 microorganisms in active surveillance in these hospitals, 67 strains were resistant to 23 % and of the 67 strains 22 (7.6%) were, Pseudomonas aeruginosa of hospitalized patients, of a German Nicaraguan public hospital and private clinics, Central Managua Hospital. The identification of the genus, species and the antimicrobial susceptibility test was carried out using the VITEK2 compact system, which was taken three UFCs of the MacConkey, for the preparation of the inoculum, then a homogeneous suspension was performed in a test tube containing 3ml of 0.45% saline solution, for the remaining cards 145µl were passed in tubes with 3ml of saline solution, DenSiCheK Plus was adjusted to the standard 0.5-6.3 of McFarland, and it was assembled (GN, AST -XN06 and AST - GN69), GN: 64 biochemical tests, AST XN06: AN, ATM, CF, CTX, CTT, FOX, CPD, CZX, CXM, DOR, MEM, MXF, NA, NOR, PIP, TE, TIC, TCC, TGC , AST GN69: AMC, AM, SAM, CZ, FEP, CAZ, CRO, CIP, ETP, GM, IPM, LEV, FT, TZP, TM, SXT. The quality control of the cards were established with the use of reference strains, Pseudomonas aeruginosa ATCC 27853, Escherichiacoli, ATCC25922, Escherichia coli ATCC35218, K. Pneumoniae ATCC700603, the suspicion of carbapenemase-producing strains was taken when Imipenem and Meropem had a minimum inhibitory concentration, Imipenem 2-4 µg / mL and for Ertapenem 2 µg / mL, a synergy test was screened with ethylenediaminetetraacetic acid (10 µg or 0.1 M), the McFarland scale 0.5 of the strains to be evaluated was prepared using the Kirby Bauer method, it was incubated 18-24hr. The Hodge Test was carried out, as a complementary method, using reference strains ATCC25922 of E. coli (Amjad A, 2011) (Cercenado, 2015). See figure 1.
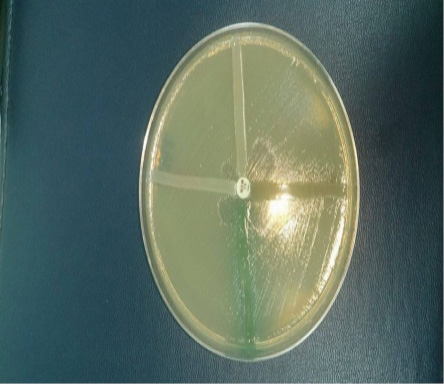
Figure 1. Hodge test
DNA extraction in Pseudomonas eruginosa. From the MacConkey agar cultivation for 18-24 hours at 37 °C, a roast of CFU was taken, inoculated into an Eppendorf tube containing 100µl of nuclease-free water, boiled in a water bath for 10 minutes, then centrifuged at 12000 rpm for 5 minutes and 80 µl of the supernatant was extracted, the concentration of DNA extracted in nanodrop lite 2763 was determined. (Armando Guevara, 2009)
The genotypic detection, which encodesb lactamase metallo, a multiplex PCR was performed, isolated from Pseudomonas aruginosa. In which IlustrapureTagReady-To-Go PCR beads were used, the concentration corresponds to Tris-HCl10mM, (pH 9.0), KCl50mM, MgCl21.5mM, 200 µM dNTPs and 2.5 µ Taq DNA polymerase. A mix of primers was made corresponding to a final volume in the 25ul beads (Edgar Gonzales-Escalante, 2011). See table 1.
|
Primers |
Sequence |
Size |
|
IMPF |
5’-GGAATAGAGTGGCTTAAYTCTC-3’ |
188 Pb. |
|
IMPR |
5’-CCAAACYACTASGTTATCT-3’ |
|
|
VIMF |
5’-GATGGTGTTTGGTCGCATA-3’ |
390 Pb. |
|
VIMR |
5’-CGAATGCGCAGCACCAG-3’ |
|
|
GIMF |
5’-TCGACACACCTTGGTCTGAA-3’ |
477 Pb. |
|
GIMR |
5’-AACTTCCAACTTTGCCATGC-3’ |
|
|
SIMF |
5’-TACAAGGGATTCGGCATCG-3’ |
570 Pb. |
|
SIMR |
5’-TAATGGCCTGTTCCCATGTG-3’ |
|
|
SPMF |
5’-AAAATCTGGGTACGCAAACG-3’ |
271 Pb. |
|
SPMR |
5’-ACATTATCCGCTGAAACAGG-3’ |
Table 1. Design of primers
Amplification Program. The following 94 °C denaturation program was used for 5 minutes; followed by 36 cycles denaturation 94 °C, for 30s; hybridization 52 °C, for 40s; amplification 72 °C, for 50s, final extension 72 °C, for 5 min, final temperature 4 °C. Samples were analyzed in a MasterCycler, Eppendorf Brand, Model number 5341 (Edgar Gonzales-Escalante, 2011).
Electrophoresis. The PCR product was evaluated on a 2% agarose gel with 0.5 µg/mL ethidium bromide, the electrophoresis was run at 120 volts for 60 minutes the DNA bands of the different genotypes were visualized in a light chamber ultraviolet and photographed. See figure 2.
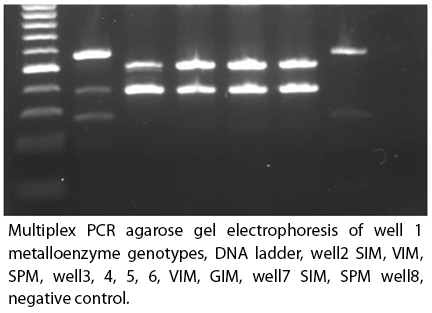
Figure 2.
Results
The universe was comprised of 288, strains of these 22 strains of Pseudomonas aeruginosa, which corresponds to 7.6%, the strains analyzed presented multi-resistance to various antibiotics analyzed, 7 (32%) strains, presented resistance to Tigecycline and none of the strains had resistance to Colistina.
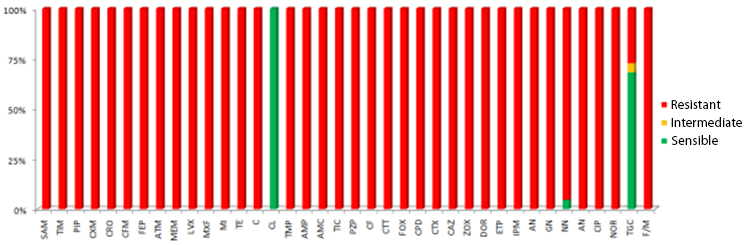
Graph 1.
Discussion
The microbial resistance has reached a critical point that the antimicrobials available for these microorganisms are very scarce, because they have a high range of mechanisms to neutralize various therapeutic options, the increase in resistant microorganism is more frequent, this complicates the situation according to Infectious process, Pseudomonas aeruginosa, constitutes one of the main causes of in-hospital mortality, according to Nicolau and Oliver, IMP and VIM, are the genes most frequently found in this type of microorganism and that at any time may be causing outbreaks, in our study We found 68% of strains carrying VIM, and combined with SPM and GIM, these genes are transmitted by horizontal transfer facilitating dissemination by plasmids and in case of SPM, it is highly related by integron, these genes are highly distributed worldwide in some countries report higher prevalence, the most worrying is that in our country, which strains with reduced sensitivity to Colistina are being detected, which is the alternative to this super bacterium capable of hydrolyzing various antibiotic families, according to Montero the main problem of Colistina, is that very limited information on its efficacy, safety, and dosage, and may cause nephrotoxicity, manifests as tubular necrosis (Carlos Juan Nicolau, 2010).
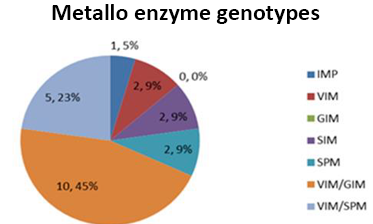
Graph 2.
The increase in resistance worldwide translates into a drastic therapeutic reduction, the alternative in our hospital services is to enforce sanitary hygienic regulations applying effective control measures to avoid nosocomial infections among various patients and apply an appropriate therapy, this would reduce the risk of mortality, economic cost and hospital stay since the risk of mortality is greater when the infectious process is a bacteremia. It was determined that 91% of the strains were concentrated in a critical area such as pediatric ICUs. (Jaime Labarca L. and Rafael Araos B., 2009).
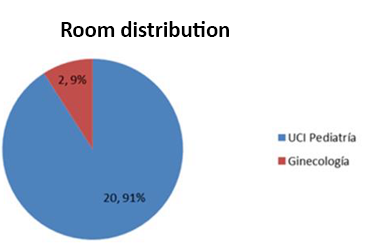
Graph 3.
Depending on the type of sample where the microorganism was isolated, its highest frequency was tracheal aspirate 7 (32%), followed by secretion 5 (23%), blood culture 5 (22%) together with a catheter tip. According to Montero, nosocomial infections by Pseudomonas aeruginosa is attributed to patients undergoing mechanical ventilation, antibiotic treatment, and post-surgical processes, Pseudomonas has the ability to colonize all parts of the body and develop various infectious processes (Carlos Juan Nicolau, 2010).
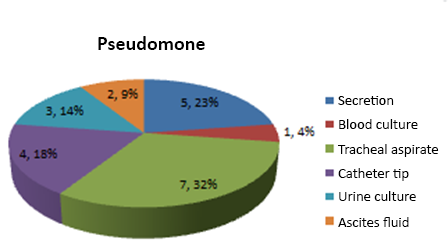
Graph 4.
Conclusion
The prevalence of Pseudomonas aeruginosa, producer of carbapenemases, in hospitals in Nicaragua is low but should be of concern for hospital authorities because the strains contain various types of genes combined as; VIM and GIM, VIM and SPM, which can trigger epidemic outbreaks, the frequency with which new strains resistant to carbapenems appears is evident, this evidence leads to the search for strategies to avoid the dispersion of multiresistant strains in different health units at the time of transfer of a carrier patient to another hospital unit.
Bibliography
Amjad A, M. I. ( December de 2011). Modified Hodge test: A simple and effective test for detection of carbapenemase production. IRAN. J. MICROBIOL, 189-193.
Armando Guevara, J. d. (2009). Detección del gen blaVIM-2 en cepas de Pseudomonas aeruginosa productoras de metalo β-lactamasa aisladas en una unidad de cuidados intensivos en Ciudad Bolívar, Venezuela. Rev Chil Infect , 26 (4)(336-341).
Carlos Juan Nicolau, A. O. (2010). Carbapenemasas en especies del género Pseudomonas. Enferm Infecc Microbiol Clin. , 19-28.
Cercenado, E. (2015). Detección de enterobacterias productoras de carbapenemasas en la rutina del laboratorio. Rev Esp Quimioter, 8-11.
Edgar Gonzales-Escalante, W. V.-T.-M.-P.-P.-G.-V.-C.-C.-S. (2011). METALO-β-LACTAMASAS EN AISLAMIENTOS CLÍNICOS DE Pseudomonas aeruginosa EN LIMA, PERÚ. Rev Peru Med Exp Salud Publica.
Jaime Labarca L. y Rafael Araos B. (2009). Resistencia antimicrobiana:Problema en aumento y soluciones escasas. Rev Chil Infect , 8-9.
Karen Bush, a. G. (2010). ANTIMICROB. AGENTS CHEMOTHER, 969–976.
M. Bodi, J. G. (2007). Pseudomonas aeruginosa: tratamiento combinado frente a monoterapia. Med Intensiva.
prevention, C. f. (s.f.). Modified Hodge test for carbapenemase detection in enterobacteriaceae. Obtenido de Available at http://www.ndhealth.gov/microlab/Uploads/HodgeTest.pdf.