ISSN 2410-5708 / e-ISSN 2313-7215
Year 9 | No. 24 | p. 138-150 | February - May 2020
© Copyright (2020). National Autonomous University of Nicaragua, Managua.
This document is under a Creative Commons
Attribution-NonCommercial-NoDerivs 4.0 International licence.
Evaluation of DNA extraction methodologies from recalcitrant plants
https://doi.org/10.5377/torreon.v9i24.9723
Submitted on February 13th, 2020 / Accepted on February 20th, 2020
B.A. Taryn Suazo Ubieta
Thesis specialists
Biotechnology Laboratory, UNAN-Managua
M.Sc. Samantha Miranda Calero
Researcher
Biotechnology Laboratory, UNAN-Managua
Keywords: DNA, extraction, commercial kits, protocols, recalcitrant
ABSTRACT
Obtention of high-quality purified deoxyribonucleic acid (DNA) is key for the subsequent application of molecular techniques during the characterization of species under study. In this regard, plant extracts can be challenging due to its complex metabolites composition and to the fact that often these chemicals concentration differs across varieties and even samples. This is, for instance, the case for recalcitrant samples of cocoa varieties (Theobroma cacao) and coffee (Coffea arabica), where DNA loss can occur during sample maceration due to phenolization. In this research, three commercial kits, already known by producing excellent recoveries for non-recalcitrant varieties, were evaluated. Additionally, the modification of traditional protocols [Doyle & Doyle (1987) and Doyle & Doyle (1990)] was also evaluated. Results obtained confirm the effect of variety and species on DNA extraction and for some species modified methods may yield DNA recovery and purity higher than those obtained by commercial kits.
INTRODUCTION
The continuous technological growth accompanied by the need to solve priority problems has encouraged the implementation of molecular techniques such as polymerase chain reaction (PCR) and sequencing. In the agricultural field, these techniques have applications that include molecular characterizations, identification of genes or mutations, among others, and in all cases, the initial and fundamental stage is the extraction of deoxyribonucleic acid (DNA) from plants of industrial, ornamental and agri-food interest, where its utility is defined by the quality and quantity obtained.
In general, the isolation of high-quality nucleic acids from the tissue of many plants is notoriously difficult (Hughes D.W. and Galau G., 1988). Inefficient cell lysis or high levels of nuclease activity have been treated in different ways, however, in multiple higher plants, the aqueous phase is affected by secondary metabolites when co-extracted together, especially in mature tissue, interfering with the isolation of biologically active nucleic acids (Baker SS, et al., 1990).
Recalcitrant plants such as cocoa (Theobroma cacao) and coffee (Coffea arabica) are rich in secondary metabolites such as phenolic compounds as well as polysaccharides, these compounds present in plant tissue can also inhibit amplification by PCR (Jobes et al., 1995; Wilson, 1997) and hinder the extraction of their DNA in the quantity and quality required. Additionally, besides the characteristics of the plants analyzed, their state of maturity, variety or cultivation of origin and type of tissue used should be considered.
There are currently various methods of DNA isolation ranging from chemical to commercial, and although DNA extraction kits promulgate the removal of most plant tissue inhibitor compounds more efficiently than “homemade” methods of isolation, experimental evidence continues finding unsatisfactory results in DNA extractions from recalcitrant matrix or with very specific biochemical characteristics (high content of polysaccharides and polyphenols, among others), compromising their quality and even their application to different molecular analyzes. It is due to this importance that the purification protocol is a fundamental criterion that must be considered for the purposes and scope of an investigation.
MATERIALS AND METHODS
Sample collection
The foliar tissue samples (leaves) used came from cocoa trees with acriolladas (native) characteristics and non-clonal coffee varieties, with intermediate maturity, medium size and in the good phytosanitary state. The leaf samples and their petiole were collected, disinfected and refrigerated until they were transferred to the Laboratory to avoid their degradation (oxidation). In the Laboratory they were characterized morphologically to corroborate their identity and were stored at -20°C properly identified until processing.
Sample Processing
Weighed 25-100 mg of tissue, and due to the high rate of oxidation of the samples, two different homogenization methods were tested: leaf drying at 45°C for 16 hours combined with maceration with calcined quartz sand adding amounts variants of polyvinylpyrrolidone (PVP), as well as the use of liquid nitrogen ensuring maceration and cold storage of the sample until incubation. It should be noted that all the micropilons used were acclimatized to prevent loss of the sample by application of heat.
Plant DNA extraction methodologies
Three commercial kits were compared: Promega Wizard SV Genomic DNA Purification System (A2360), Roche High Pure PCR Template Preparation Kit (11 796 828 001), and Qiagen DNeasy Plant Mini Kit (69104). Additionally, the results obtained from the modification to the chemical extraction protocols described by Doyle & Doyle (1987) and Doyle (1990) were evaluated because when implementing the original protocols no DNA was obtained from the samples. All tests with chemical protocols were performed in duplicate, with negative control. Grass samples (Pennisetum purpureum) and neem (Azadirachta indica) were included as positive extraction controls in the tests where it was considered relevant due to the spectrum of the kit or protocol.
Extractions with Promega (A2360), Roche (11 796 828 001) and Qiagen (69104) kits were performed according to the protocol established by the manufacturers. The extraction carried out according to the modified protocol of Doyle and Doyle (1987) started from 100 mg of foliar tissue and 30 mg of purified sea sand, being macerated with 1000 μL of hexadecyltrimethylammonium bromide extraction buffer (CTAB).) 2X preheated to 65°C. Once macerated, it was incubated at room temperature for 5 minutes and then at -20°C for 12 minutes. 20 µL of RNase A was added and incubated for 20 minutes at 37°C. Subsequently, 10 μL of proteinase K (13 mg / mL) was added, incubating again for 20 minutes at 60°C and 7 minutes on ice. Three washes were performed with 600 μL of phenol: chloroform: isoamyl alcohol (25: 24: 1). Then, 50 µL of 10M ammonium acetate was added and incubated at -20°C after the addition of 500 µL isopropanol (4°C). The supernatant was decanted after centrifugation and 1 mL of 70% ethanol (-20 °C) was added, mixed by inversion, placed at rest for 5 minutes at room temperature and centrifuged again to obtain the precipitate that was dried for 40 minutes. Finally, the DNA was rehydrated with 200 μL of 1X TE buffer.
The Doyle protocol (1990) with modifications by Shepherd & McLay (2011) and Tibbits, McMannus, Spokevicius, & Bossinger (2006) was implemented starting from 50 mg of foliar tissue macerated with liquid nitrogen. The modifications made include the homogenization of the maximum speed stirring times in Vortex for 1 minute, also, 500 μL of CTAB buffer previously heated to 65°C was added independently, followed by the 99.9% β-mercaptoethanol used, proteinase K (13 mg/mL) and the mixture of NaCl: BSA (5: 1). The samples were incubated for 20 h at 65°C mixing in Vortex for 1 minute during and after incubation. Subsequently, isopropanol was added and mixed by inversion, centrifuged and incubated at -20°C and in the first resuspension step in 10 mM Tris-HCl was stored at 4°C for 16 h. It should be noted that after this period of rest, 2.4 M sodium acetate and absolute ethanol were added, mixing by inversion followed by centrifugation, washing with ethanol, drying the pellet and resuspension in TE until stored at 4°C.
As a final test of this modified protocol, 50 mg of leaf tissue from a sample of cocoa (two varieties) and coffee (one cultivar) were added by adding a lower concentration of β-mercaptoethanol (10%) and a third wash with chloroform was added to obtain cleaner samples, since the elimination of secondary metabolites that could interfere with the amplification of the samples is necessary using the polymerase chain reaction (PCR) technique.
Quality assessment of extracted plant DNA
The quality of the DNA obtained was verified by agarose gel electrophoresis with a concentration of 0.8% and ethidium bromide staining. To load the samples into the gel, these were mixed with 6X loading dye (Thermo Fisher Scientific, R0611) and a 100 bp molecular weight marker standard (Fisher Scientific, BP2573-100) was used. The gels were run at 80V for one hour and were visualized in an ultraviolet transilluminator model BBA (Fisher Scientific).
The quantification of the double-stranded DNA concentration (dsDNA) was performed by determining its absorbance at 260 nm in the Cary 50-BIO spectrophotometer (Varian). The purity of the samples was determined by quantifying the presence of proteins at 280 nm and calculating the A260 / A280 ratio; the turbidity correction was also performed at 320 nm.
Finally, the samples with the best quality and purity results were amplified by polymerase chain reaction (PCR) using microsatellite molecular markers (SSR) and verified on an agarose gel to 1.8%. During the PCR, 25 µL reaction volumes containing 1 µL of DNA, 1X of Go Taq® Colorless Master Mix (M7133) and 1 µM of each first (Eurofins Genomics) were used. The amplification program consisted of the initial denaturation (94°C) for 3 minutes (min.), Followed by 30 cycles of 94°C for 30 seconds (s), 30 s alignment temperature of each first and 45 s at 72 °C, with a final extension at 72 °C for 3 min. The amplification products were analyzed by electrophoresis in 1.8% Low EEO/Multipurpose agarose gel (BP160-500) stained with ethidium bromide (10 mg / mL).
RESULTS AND DISCUSSION
Promega Wizard SV Genomic DNA Purification System Kit (A2360)
50 and 100 mg of both fresh cocoa leaves were used, pre-dried in the oven and macerated with liquid nitrogen, however, bands were not visualized on the agarose gel (0.8%), obtaining poor quality results (Figure 1).
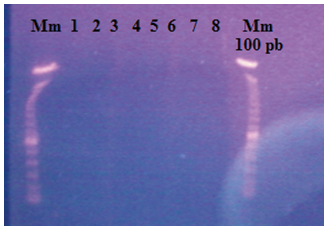
Figure 1. Extraction results with Promega kit (A2360) in agarose gel (0.8%). Mm: 100 bp molecular weight marker, lines 1 and 2: 50 mg of pre-dried and macerated sample with sand, lines 3 and 4: 100 mg of pre-dried and macerated sample with sand, lines 5 and 6: 50 mg of sample macerated with liquid nitrogen, lines 7 and 8: 100 mg of sample macerated with liquid nitrogen.
Roche High Pure PCR Template Preparation Kit (11 796 828 001)
Extraction was performed using from 25 to 45 mg of cocoa and grass samples (control) stored at -20°C and macerated using sand or liquid nitrogen. Minimum quality results were obtained for grass and null samples for cocoa (Figure 2).

Figure 2. Extraction results with Roche kit (11 796 828 001) in agarose gel (0.8%). 100 bp: molecular weight marker, line 1: 25 mg of grass macerated with liquid nitrogen, line 2: 25 mg of grass pre-dried and macerated with sand, line 3: 45 mg of grass macerated with liquid nitrogen, line 4: 45 mg of grass pre-dried and macerated with sand, lines 5: 25 mg of cocoa macerated with liquid nitrogen, line 6: 25 mg of cocoa pre-dried and macerated with sand, line 7: 45 mg of cocoa macerated with liquid nitrogen, line 4: 45 mg of cocoa pre-dried and macerated with sand.
Qiagen DNeasy Plant Mini Kit (69104)
The extraction was made from 100 mg of cocoa leaf tissue from 3 different trees macerated with 30 mg of purified sea sand. The samples of genomic DNA extracted were analyzed after visualization on 0.8% agarose gel where very low recovery was found for samples 1 and 3, and greater band intensity in sample 2 (Figure 3). This may be due to differences in the content of compounds that interfere with DNA recovery depending on the variety under study.
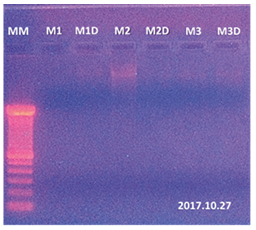
Figure 3. Extraction results with Qiagen kit (69104) in agarose gel (0.8%). MM: 100 bp molecular weight marker, M1 lines: 100 mg of cocoa sample 1, M1D line: M1 sample replica, M2 line: 100 mg of cocoa sample 2, M2D line: M2 sample replica, M3 line: 100 mg of cocoa sample 3, M3D line: M3 sample replica.
Protocol Doyle and Doyle (1987) modified
In this protocol, no positive results were obtained in the extraction, discarding this protocol for the varieties under study (Figure 4).

Figure 4. Extraction results with Doyle and Doyle protocol (1987) on agarose gel (0.8%). MM: 100 bp molecular weight marker, M1 lines: 100 mg of cocoa sample 1, M1D line: M1 sample replica, M2 line: 100 mg of cocoa sample 2, M2D line: M2 sample replica, CN line: negative control of the medium.
Doyle Protocol (1990) with modifications by Shepherd & McLay (2011) and Tibbits, McMannus, Spokevicius, & Bossinger (2006)
Genomic DNA extraction from samples of cocoa, grass and neem leaf tissue with its duplicate was performed using 50 mg of leaf tissue, (Figure 5). In the gel, the need for additional washes can be evidenced, which constituted the final modification of the protocol (figure 6). The results obtained from the final modification of the Doyle protocol (1990) showed better DNA recoveries in addition to an appropriate quality for their characterization.
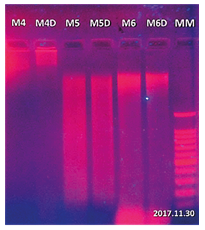
Figure 5. Results of extraction with Doyle protocol (1990) in agarose gel (0.8%). MM: 100 bp molecular weight marker, M4 lines: 50 mg of cocoa sample, M4D line: M4 sample replica, M5 line: 50 mg grass sample, M5D line: M5 sample replica, M6 line: 50 mg of neem sample, M6D line: a replica of the M6 sample.
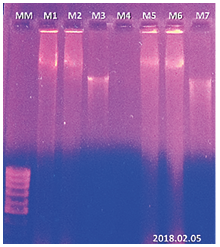
Figure 6. Results of the last test with the modified agarose gel protocol (0.8%). MM: 100 bp molecular weight marker, line M1: 50 mg of cocoa sample, line M2: replica of sample M1, line M3: 50 mg of coffee sample, line M4: negative control, line M5: 50 mg of cocoa sample, line M6: replica of sample M5, M7: 50 mg of coffee sample.
Quantification of the dsDNA
The amount of DNA obtained for the five extraction methodologies is variable, both by the method and by a matrix (figure 7). In all cases, coffee samples showed better recovery than cocoa samples. Regarding the methodology, the highest amount of DNA (0.99 mg/ml in coffee and 0.70 mg/ml in cocoa) was determined in the modified Doyle (1990) protocol, followed by the Qiagen kit (0.53 mg/ml in coffee and 0.34 mg/ml in cocoa) and the Promega kit (0.24 mg/ml in coffee and 0.17 mg/ml in cocoa).
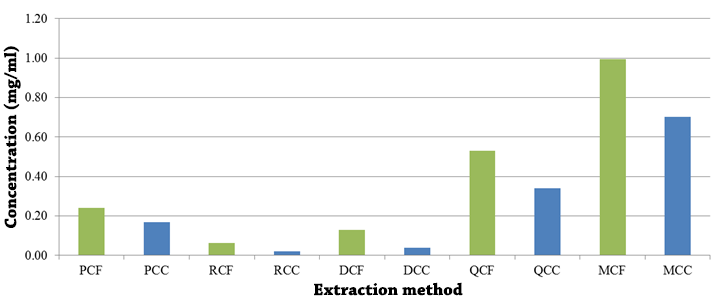
Figure 7. Concentration of DNA (mg/ml) from coffee (CF) and cocoa (CC) cultivars obtained through commercial kits: Promega (P), Roche (R), Qiagen (Q) and chemical methods: Doyle & Doyle (1987) (D) and Doyle (1990) with modifications by Shepherd & McLay (2011) and Tibbits, McMannus, Spokevicius, & Bossinger (2006) (M). PCF: Promega method and coffee samples, PCC: Promega method and cocoa samples, RCF: Roche method and coffee samples, RCC: Roche method and cocoa samples, DCF: Doyle & Doyle method (1987) and coffee samples, DCC: Doyle & Doyle method (1987) and cocoa samples, QCF: Qiagen method and coffee samples, QCC: Qiagen method and cocoa samples, MCF: Modified Doyle method (1990) and coffee samples, MCC: Doyle method ( 1990) modified and cocoa samples.
DNA purity
Similarly, the purity of the samples is variable in terms of the methodology used, although it should be noted that it does not show the same variation observed in the quantification of the DNA concerning the matrix (figure 8).
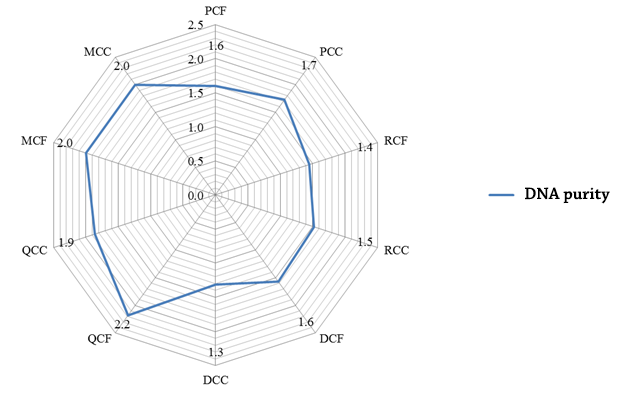
Figure 8. DNA purity (mg/ml) of coffee (CF) and cocoa (CC) cultivars obtained by commercial kits: Promega (P), Roche (R), Qiagen (Q) and chemical methods: Doyle & Doyle ( 1987) (D) and Doyle (1990) with modifications by Shepherd & McLay (2011) and Tibbits, McMannus, Spokevicius, & Bossinger (2006) (M).
In general, the results obtained both in the integrity of the samples by electrophoresis as well as the concentration and purity of the extracted DNA coincide with the drawbacks described by Li et al. (2002) and Sharma et al. (2002), which describe the problem related to the co-extraction of polysaccharides and polyphenols that act as inhibitors during DNA extraction, and which, depending on their concentration, can oxidize the DNA during the lysis process. Sharna et al. (2002) describe that even closely related species may require different DNA isolation protocols, which coincides with the experience before this test where it has been possible to recover genetic material with the Qiagen, Promega kit and the methods of Doyle & Doyle (1987) for certain cocoa varieties. It should be noted that cocoa samples, which reflected the lowest concentration in all cases, are considered acrylate by their productive, morphological characteristics, in addition to their aroma and flavor.
Likewise, López P., López A. and Marulanda M. (2011), report in their standardization of the DNA extraction of Tabebuia rosea and Cordia alliodora, that through Qiagen’s Plant DNeasy mini kit, it was not possible to extract and amplify the T. rosea samples because the kit requires young tissues or that contain low levels of polyphenols (Li et al. 2002), the opposite case of the selected samples as indicated by Martínez (2010) and Arbeláez et al. (2011) present various secondary metabolites. This type of sample matches the chemical characteristics of cocoa and coffee.
Amplification using the best quality DNA
After verification of the quality of the extracted DNA, the sample of coffee obtained using the modified Doyle protocol (1990) was selected as the best result and amplified with 10 microsatellite markers (SSR) internationally described for the molecular characterization of coffee. The results obtained were satisfactory in terms of clarity and reproducibility in the bands (figure 9).
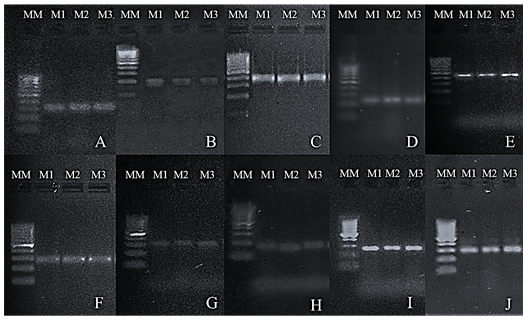
Figure 9. PCR products obtained from the amplification of coffee DNA extracted by the Doyle modified protocol (1990) using 10 pairs of markers on Low EEO / Multipurpose agarose gel (1.8%). MM: 100 bp molecular weight marker. M1, M2, and M3: replicas of the samples. (A) Amplicons with the CarM096 marker. (B) Amplicons with the CarM101 marker. (C) Amplicons with the CarM049 marker. (D) Amplicons with the CarM069 marker. (E) Amplicons with the CarM092 marker. (F) Amplicons with the CarM105 marker. (G) Amplicons with the M20 marker. (H) Amplicons with the M24 marker. (I) Amplicons with the CarM051 marker. (J) Amplicons with the CarM050 marker.
CONCLUSIONS
The quality and quantity of the extracted DNA are influenced by the methodology used as well as by the nature of the matrix under study. Of the commercial methods used, the best results in terms of quantity and purity were obtained by extraction with the Qiagen kit, although it should be noted that the results were low recovery. As for the protocols evaluated, Doyle (1990) with modifications by Shepherd & McLay (2011) and Tibbits, McMannus, Spokevicius, & Bossinger (2006) presented the best recoveries and purity, even higher than the commercial kit. It should be noted that this behavior cannot be generalized to all plant varieties, even of the same species, because through the Promega and Qiagen kits, as with the Doyle and Doyle (1987) methodology, good recoveries of DNA for other varieties of cocoa and coffee demonstrating the influence of the variety of species from which the tissue to be analyzed is obtained.
THANKS
This study was carried out with the support of the funding received by the Fund for Research Projects (FPI as in Spanish) of UNAN-Managua.
BIBLIOGRAPHIC REFERENCES
Alejos, L., Aragón, M., & Cornejo, A. (2008). Extracción y purificación de ADN. México: UNAM.
Arbeláez, J., Acevedo, J. y Jaramillo, R. (2011). Metabolitos secundarios en el guayacán amarillo y en el guayacán rosado. Scientia et Technica 47:297- 301.
Baker S.S., Rugh C.L. y Kamalay J.C. (1990). RNA and DNA isolation from recalcitrant plant tissues. Biotechniques; 9(3):268-72.
Doyle, J. (1990). DNA PROTOCOLS FOR PLANTS. NATO ASY Series, 283-284.
Doyle, J., & Doyle, J. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical bulletin, 19:11–15.
Hughes D.W. y Galau G. (1988). Preparation of RNA from cotton leaves and pollen. Plant Molecular Biology Rep. 6:253-257.
Jobes D. V., Hurley D. L. y Thien L.B. (1995). Plant DNA isolation: A method to effi ciently remove polyphenolics, polysaccharides, and RNA. Taxon 44: 379 – 386.
Li, Y. X., Su, Z. X. y Chen, F. (2002). Rapid Extraction of Genomic DNA From Leaves and Bracts of Dove Tree (Davidia involucrata). Plant Molecular Biology Reporter 20(2):185.
Lopez Mora, P., Lopez Gutierrez, A., & Marulanda-Angel, M. (2011). Estandarización de la extracción de adn genómico en Tabebuia rosea (Bertol.) DC. y Cordia alliodora (Ruiz y Pav.) Okén. Temas Agrarios, 16(2), 28-41.
Martínez, A.J. (2010). Principios activos en el Guayacán Amarillo (Tabebuia chrysantha Jacq. Nicholson, Bignoniaceae) y en el Guayacán Rosado (Tabebuia rosea Bertol. DC). Proyecto de Grado, Universidad del Quindío, Armenia - Colombia.
McLay, T. (4 de Septiembre de 2017). Hight quality DNA extraction protocol from recalcitrant plant tissues. Obtenido de protocols.io.
QIAGEN. (2006). Protocolo: Purification of total DNA from Plant Tissue (Miniprotocol). Estados Unidos: QIAGEN Group.
QIAGEN. (2011). DNeasy® Plant Mini Kit . Estados Unidos: QIAGEN Group.
Sharma, A.D., Gill, P.K. y Singhdna, P. (2002). Isolation From Dry and Fresh Samples of Polysaccharide-Rich Plants. Plant Molecular Biology Reporter 20(4):415.
Shepherd, L., & McLay, T. (2011). Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue. Journal of Plant Research, 311-314.
Tibbits, J., McMannus, L., Spokevicius, A., & Bossinger, G. (2006). A rapid method for tissue collection and high-throughput isolation of genomic DNA from mature trees. Plant Molecular Biology Reporter, 81-91.
Wilson, I. G. (1997). Inhibition and facilitation of nucleic acid amplification. Applied and Environmental Microbiology 63: 3741 – 3751.